The GRC sequences thousands of microbial samples each year. The high throughput of our sequencers and the small genome sizes of these samples means that a lot of libraries are needed to keep our sequencers running at full capacity. Automation of library preps is key to keeping instruments busy, reducing error, and maximizing the productivity of our lab staff.
We have used automation to prepare large batches of libraries for Illumina sequencing for several years. Earlier this year, we began testing preparation of long-insert libraries for our PacBio RS II platform.
Our Biomek FXP (BeckmanCoulter) is a Dual Multi-channel Span-8 with an integrated thermal cycler. The combination of a 96-channel pipetting head and Span-8 pipetting head (which has eight independently controlled channels) allows for the preparation of up to 96 libraries at a time and the minimization of master mix dead volumes.
A script for preparing SMRTbell libraries developed by Todd Hartley at NCI/SAIC-Frederick was downloaded from Pacific Biosciences SMRT Community. We modified to the protocol to accommodate the specific tips and reagents tubes our lab uses, and to optimize reaction mixing.
Deck Layout
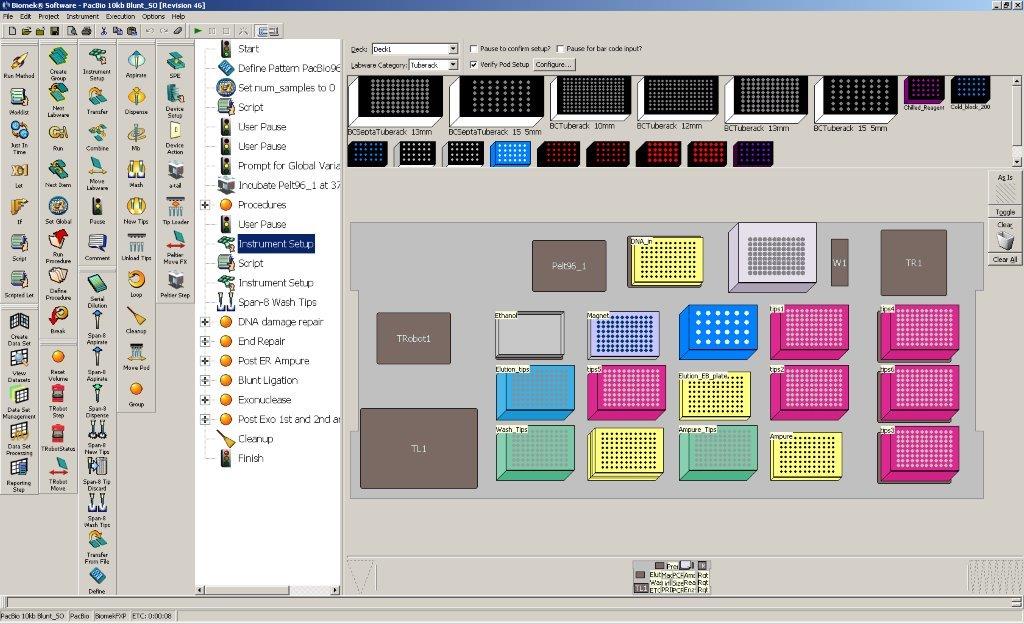
Prior to being loaded on the Biomek, samples are sheared with g-TUBEs (Covaris, Woburn, MA), targeting an average fragment size of 20kb. Master mixes for each reaction are prepared and placed in the robot. The following steps are performed by the Biomek:
- DNA damage repair
- End Repair
- Ampure clean up
- SMRTbell Ligation
- Exonuclease
- Ampure clean up
Once the run is complete, the libraries are removed and size-selection is performed using the BluePippin (Sage Science, Beverly, MA).
| |
manual preps |
robotic preps |
| Number of libraries (n) |
91 |
27 |
| Average input amount (ng of sheared gDNA) |
4945 |
4951 |
| Average library size (bp) |
18507 |
18910 |
| Average library concentration (nM) |
5.1 |
3.5 |
| Average recovery (ng) |
902.8 |
707.4 |
| Average recovery (%) |
18.7 |
14.2 |
Above are data comparing libraries prepped manually and on the Biomek, from March 2014 to date. While the yields from the Biomek preps are slightly lower than manual preps, the yield is comparable and sufficient for sequencing multiple SMRT cells per library.
For more information on our full range of sequencing and analysis services, visit our Laboratory Services and Analysis Services pages. Please contact us if you have any questions.



