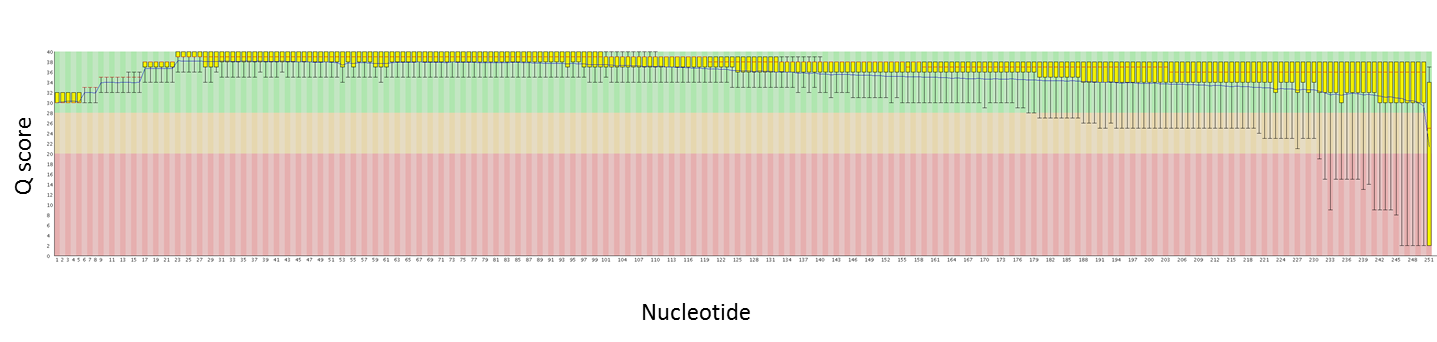
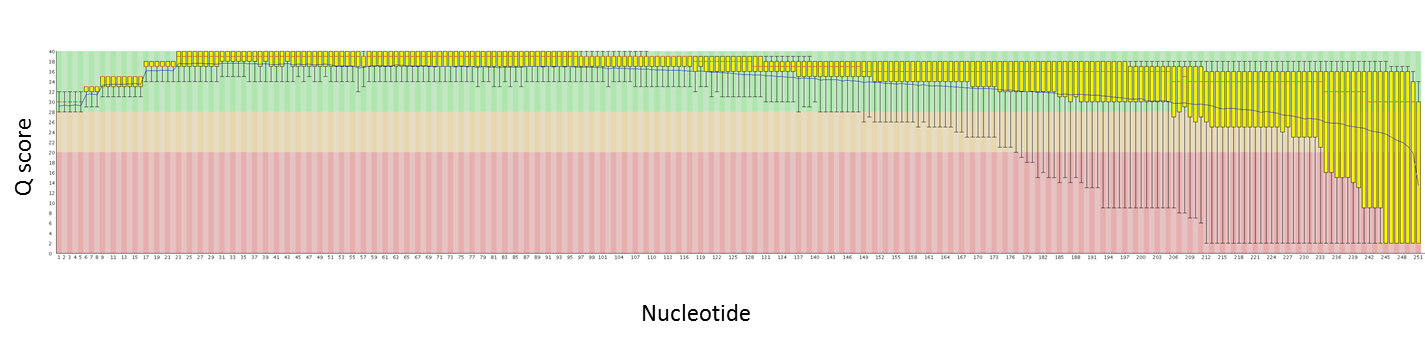
Over the past couple of months we have been evaluating the MiSeq upgrade. This upgrade includes the ability to sequence longer reads (250nt from each end, so 500 nt per library fragment) and to collect data from more clusters (both the top and bottom of each channel are imaged). We just had a 250 PE run that exceeded 30 million reads – that is just over 8 Gbases of data! This is a nice jump up from the ~13M reads (~2 Gbases) per run we were getting before.
Disclaimer: We are still in data-gathering mode to determine what the average expectation should be for each run- it would be nice to get 30 million reads from every run, but that may not happen.
Here are quality plots of a 250 PE run with genomic PE libraries. We are working to maximize quality as the read lengths increase.
Read 1:
Read 2:
We can now combine the benefits of Illumina’s high read counts with the benefits of longer reads.
Up next is to see how MiSeq/Pac Bio hybrid assemblies measure up to HiSeq/454 hybrid assemblies, and a comparison of assemblies using HiSeq 100bp PE reads vs MiSeq 250 bp PE reads.