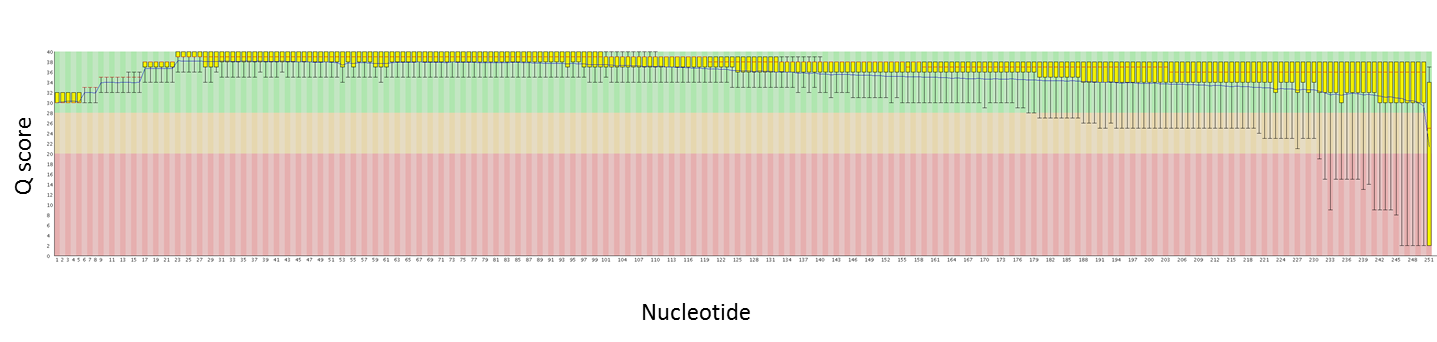
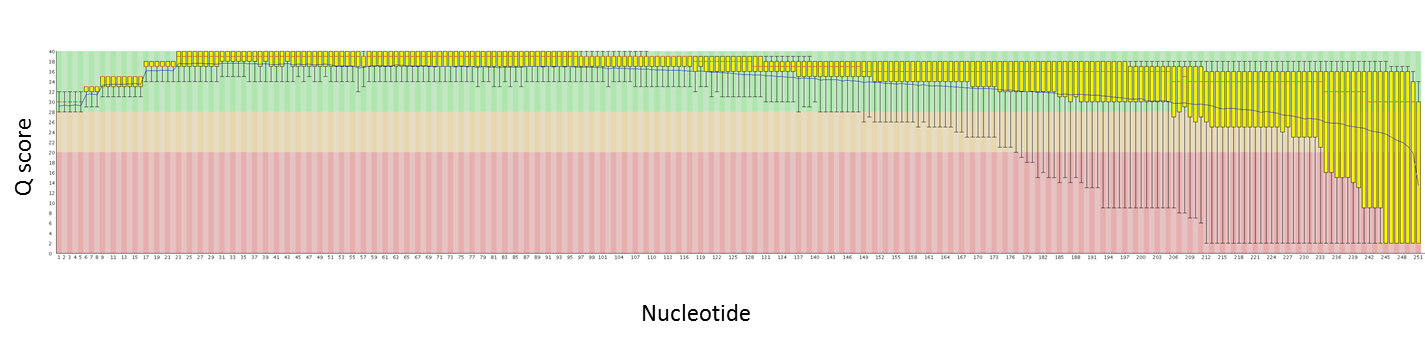
The PacBio was recently upgraded to version 1.3.3. With this upgrade comes the ability to use the XL versions of the DNA/Polymerase Binding and DNA Sequencing kits. These new kits should result in a longer average readlength (5000 bp) in comparison to the ~3000 bp average we get with the current C2 chemistry.
Using both new kits together does come at a cost. The data produced with the DNA Sequencing Kit XL 1.0 will be of a lower quality than with C2, and is recommended only when the data will be error corrected with shorter, more accurate reads.
For a boost in average read length without sacrificing quality of the reads, the DNA/Polymerase Binding Kit XL 1.0 can be used with the C2 sequencing chemistry rather than with the newer XL sequencing kit.
More details to follow…