LGTSeek will be made available as a graphical user interface (GUI) in the form of a Docker image. This saves users from having to download and install a complex pipeline and all the necessary dependencies. Users are not required to know how to use a UNIX-based environment.
An beta version of the LGTSeek Docker image is available below.
Four canned pipelines will be made available according to the four most common use cases.
PIPELINE 1
PREREQUISITES: good donor reference genome + good LGT-free recipient reference genome.
PIPELINE 2
PREREQUISITES: good donor reference genome + good recipient reference genome with LGT
PIPELINE 3
PREREQUISITES: no good donor reference genome + good LGT-free recipient reference genome
PIPELINE 4
PREREQUISITES: good donor reference genome + no good recipient reference genome
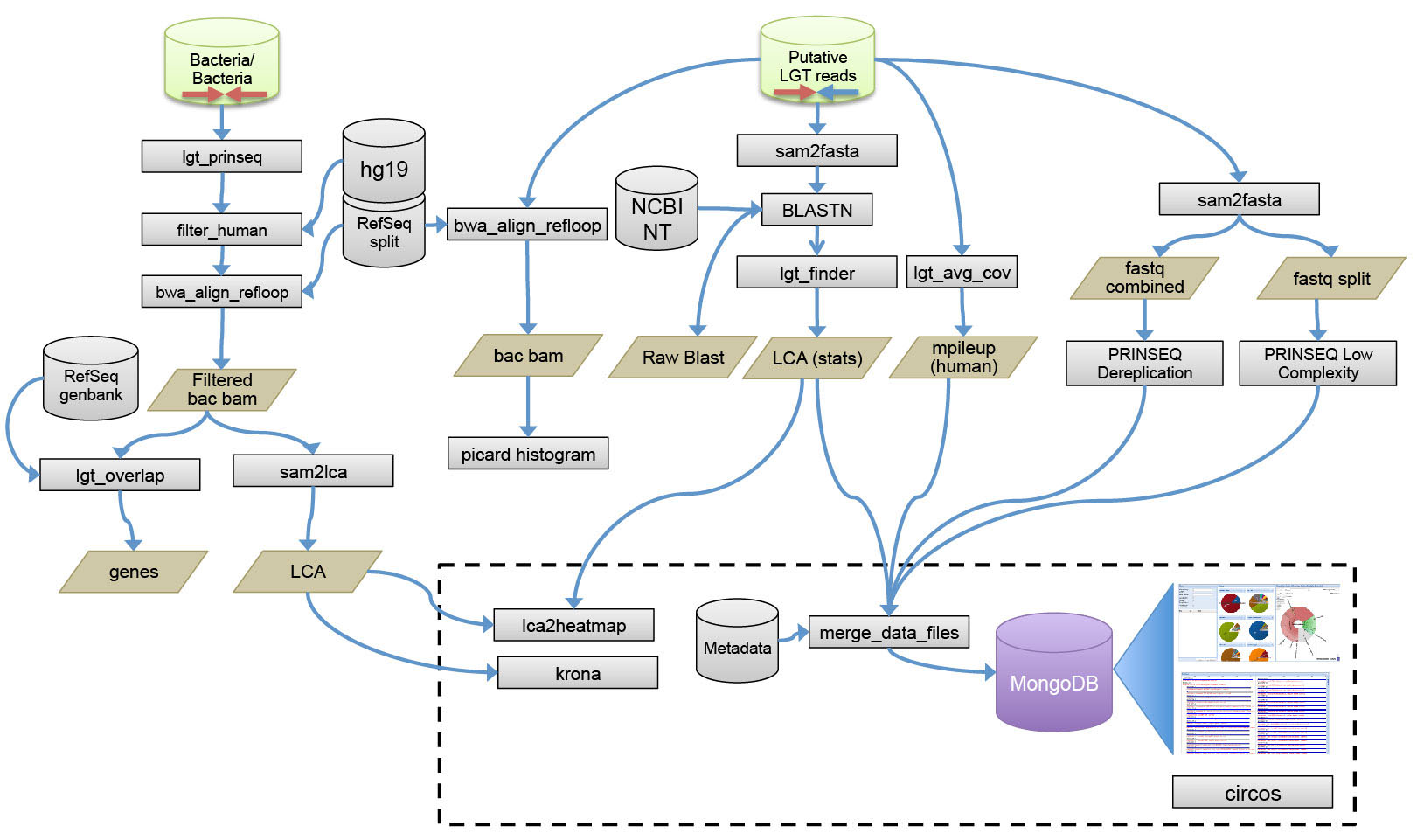
Figure1. Detailed schematic of method employed to identify putative LGT reads. Following the identification of putative LGT reads and microbiome reads with BWA, a series of steps are undertaken to remove low complexity sequences, remove duplicates, remap the reads, and generate data for the visualization interfaces. This data includes the LCA assignment, coverage, and overlaps with genes as well as Krona plots and heat maps. Where possible, existing tools are used like BWA, BLAST, MPILEUP, and PRINSEQ1.
REFERENCES
- Riley, D. R., Sieber, K. B., Robinson, K. M., White, J. R., Ganesan, A., Nourbakhsh, S., & Dunning Hotopp, J. C. (2013). Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput Biol, 9(6), e1003107.
An beta version of the LGTSeek docker image can be found here.
The code for the beta version of LGTSeek is available on GitHub here.
The code for the LGTSeek pipeline is available on GitHub here.
A tutorial for this version of LGTSeek can be found here.